43 maxquant label free quantification
MaxQuant and MSstats for the analysis of label-free data In this training we will cover the full analysis workflow from label-free, data dependent acquisition (DDA) raw data to statistical results. We'll use two popular quantitative proteomics software: MaxQuant and MSstats. MaxQuant allows protein identification and quantification for many different kinds of proteomics data (Cox and Mann 2008). maxquant:manual:beginner [MaxQuant documentation] The descriptions and screenshots in this tutorial refer to the MaxQuant GUI around version 1.4.3.14 from August 2014. ... and the "Multiplicity" will be 1 for label-free quantification, 2 if you have light and heavy labels, and 3 if you have light, medium, and heavy. Any number of labels can be checked in the lists. In this example, the ...
comprehensive evaluation of popular proteomics software workflows for ... For instance, in , three different software platforms, Progenesis, MaxQuant and Proteios ... Label-free quantification with PEAKS Q was used. PEAKS was allowed to autodetect the reference sample and automatically align the sample runs. To allow the exporting of complete results, protein significance filter was set to 0, protein fold change ...

Maxquant label free quantification
MQSS 2019 | L4: Label free quantification | Christoph Wichmann To address the drawbacks and make use of the advantages of the label-free approach, MaxQuant contains a label-free quantification module called MaxLFQ. It is a series of algorithms that are based... Comparative evaluation of label-free quantification strategies The selection of a data processing method for use in mass spectrometry-based label-free proteome quantification contributes significantly to its accuracy and precision. In this study, we comprehensively evaluated 7 commonly-used label-free quantification methods (MaxQuant-Spectrum count, MaxQuant-iB … MaxQuant error at label free normalization step - Google Groups Then no issue with maxquant search. Good luck ... I am having a similar problem, but under my description it just says the file folder and "label free_normalization 0 Label free_normalization" and then it cuts off. I'm searching 180 files and it keeps happening. All files open fine in xcalibur, and most have searched fine in smaller searches.
Maxquant label free quantification. Label-free quantification with FDR-controlled match-between-runs Missing values weaken the power of label-free quantitative proteomic experiments to uncover true quantitative differences between biological samples or experimental conditions. Match-between-runs (MBR) has become a common approach to mitigate the missing value problem, where peptides identified by tandem mass spectra in one run are transferred to another by inference based on m/z, charge state ... Comparative evaluation of label-free quantification strategies To compare the performance of widely used or recently developed label-free quantification strategies, we analyzed the data using seven distinct label-free quantification strategies ( Fig. 1 and Supplementary Table S1). MQ-SC and StPeter were based on spectral count, while other strategies were based on ion intensity. How do I quantify the label free peptides across the samples from LTQ ... For the quantification, or you do it manually (if you have a lot of free time) or you might want to use the new feature of Skyline, which now does also label free quantification Cite 1... Comparative Evaluation of MaxQuant and Proteome Discoverer ... May 26, 2021 ... MS1-based label-free quantification can compare precursor ion peaks across runs, allowing reproducible protein measurements.
The MaxQuant computational platform for mass spectrometry ... - Nature Overview of the preconfigured label-free and label-based quantification methods supported by MaxQuant. ... To enable LFQ go to the 'Label free quantification' page and select the 'LFQ' option. 28. PDF Label-free Quantification with PEAKS - Bioinformatics Solutions Inc. Label-free Quantification Protein abundances across a large number of samples can be compared via label-free quantitative method. Compared to label-based approaches, label-free quantification workflows are simpler, since no isotopic labeling is involved. In addition, less complicated data analysis is required. Label Free Quantitation using Maxquant LFQ (MaxLFQ) MaxLFQ is a generic label-free quantification technology that is readily applicable to many biological questions; it is compatible with standard statistical analysis workflows, and it has been validated in many and diverse biological projects. Our algorithms can handle very large experiments of 500+ samples in a manageable computing time. Label-Free Quantification - an overview | ScienceDirect Topics 3.7.3 Label-free quantification. Label-free quantification is a method in MS that determines the relative amount of proteins in two or more biological samples, but unlike other quantitative methods, is does not use a stable isotope that chemically binds and labels the protein. 70 Typically, peptide signals are detected at the MS1 level, and ...
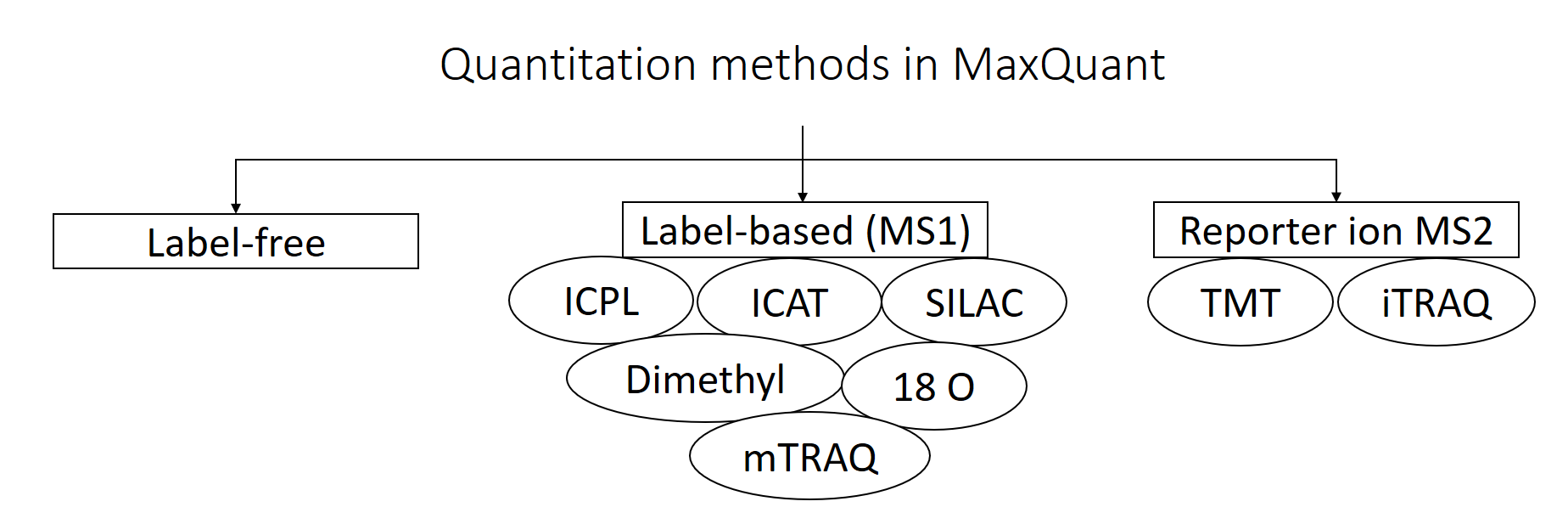
Label-free data analysis using MaxQuant - Galaxy Training Network MaxQuant offers highly accurate functionalities for many different proteomics quantification strategies, e.g. label-free, SILAC, TMT. Blood is a commonly used biofluid for diagnostic procedures. The cell-free liquid blood portion is called plasma and after coagulation serum. › doi › 10Lysosomal enzyme trafficking factor LYSET enables nutritional ... Sep 08, 2022 · Cell lines used in most experiments were MIA PaCa-2, HEK 293T and MEFs. Cell culture experiments in different nutrient conditions were performed with amino acid-free, glucose-free DMEM/F-12 reconstituted with glucose and all amino acids except leucine at standard DMEM/F12 concentrations. What is the best experimental design in Max Quant for Label Free? I am using MaxQuant to analyze some label-free experiment data. I did an affinity chromatography experiment and I have a technical triplicate of the elution and a technical triplicate of the... Label-free data analysis using MaxQuant - Galaxy Training Network MaxQuant supports different "Quantitation Methods". The three main categories are label-free quantification, label-based quantification and reporter ion MS2 quantification. In this tutorial we have chosen label-free because we did not apply any specific labeling/quantitation strategy to the samples.
Label-free data analysis using MaxQuant - Galaxy Training! May 31, 2021 ... MaxQuant supports different “Quantitation Methods”. The three main categories are label-free quantification , label-based quantification and ...
MaxQuant and MSstats for the analysis of label-free data - Lazarus In this training we will cover the full analysis workflow from label-free, data dependent acquisition (DDA) raw data to statistical results. We'll use two popular quantitative proteomics software: MaxQuant and MSstats. MaxQuant allows protein identification and quantification for many different kinds of proteomics data (Cox and Mann 2008).
Accurate Proteome-wide Label-free Quantification by Delayed ... MaxLFQ is a generic label-free quantification technology that is readily applicable to many biological questions; it is compatible with standard statistical analysis workflows, and it has been validated in many and diverse biological projects. Our algorithms can handle very large experiments of 500+ samples in a manageable computing time.
Efficient quantitative comparisons of plasma proteomes using label-free ... Select "Label free quantification" tab. If more than one parameter group is used, and LFQ normalizations are to be kept separate (see Note 3), enable "Separate LFQ in parameter groups". Keep default "Stabilize large LFQ ratios", "Require MS/MS for LFQ comparisons" and "Advanced site intensities" settings enabled.
LFQ-Analyst: An Easy-To-Use Interactive Web Platform To Analyze ... Oct 28, 2019 ... Relative label-free quantification (LFQ) of shotgun proteomics data ... to analyze and visualize MaxQuant data derived from label-free, ...
MaxQuant label-free quantification - Google Groups MaxQuant label-free quantification. 45 views. ... When I use MaxQuant for the LFQ analysis. I found some proteins from my experimental sample have high intensities and several peptides. Their intensities are 0 in the control sample. I am very interested in those proteins. But their LFQ intensities are 0 in both samples.
Comparative evaluation of label-free quantification strategies Mar 20, 2020 ... A comprehensive evaluation of 7 commonly-used label-free quantification methods was conducted. •. MaxQuant in MaxLFQ mode outperformed other ...
Plant Phosphopeptide Identification and Label-Free Quantification by ... Phosphopeptide(s) identification and label-free quantification can determine dynamic changes of phosphorylation events in plants. Both MaxQuant and Proteome Discoverer are professional software tools used to identify and quantify large-scale MS-based phosphoproteomic data.
PDF MaxQuant Information and Tutorial - uni-greifswald absolute quantification: copy numbers or concentration of protein within a sample (challenge: different peptide species with various ionization efficiencies have to be quantitatively compared within the same sample) both: label-based (relative: e.g. SILAC, ICAT, TMT; absolute: e.g. AQUA) or label-free possible 1.4.2 label-free quantification
MaxQuant MaxQuant MaxQuant is a quantitative proteomics software package designed for analyzing large mass-spectrometric data sets. It is specifically aimed at high-resolution MS data. Several labeling techniques as well as label-free quantification are supported. MaxQuant is freely available and can be downloaded from this site.
PDF Protein Groups Output (MaxQuant or MQ) - Emory University Normalization is used in the MaxQuant LFQ algorithm for label-free quantification. Firstly, the intensities for a peptide are summed up over the factions of one sample with introducing the normalization factors as unknown variables.
› yanzhi123 › p蛋白组学定量值得比较说明 - 原生態 - 博客园 Oct 21, 2019 · 我们使用Maxquant做Label Free蛋白质组学定量分析的时候,在Maxquant的参数设置时,会遇到两个参数,LFQ和iBAQ,那么,选择哪个好呢? 如果你都选上,在最终的proteingroups.txt中,会出现三列:Intensity、IBAQ、LFQ intensity,这三列中的数字,也就是蛋白的定量强度,并不 ...
Quantitative proteomics: label-free quantitation of proteins MaxQuant employs the MaxLFQ algorithm for label-free quantitation (LFQ). Quantification will be performed using razor and unique peptides, including those modified by acetylation (protein N-terminal), oxidation (Met) and deamidation (NQ). 2. Quality Control of MaxQuant Search(optional)
MaxQuant - Research Computing Documentation - University of South Florida Description. From the MaxQuant home page: MaxQuant is a quantitative proteomics software package designed for analyzing large mass-spectrometric data sets. It is specifically aimed at high-resolution MS data. Several labeling techniques as well as label-free quantification are supported. The search engine andromeda is integrated into MaxQuant ...
MaxQuant is a quantitative proteomics software package designed for analyzing large mass-spectrometric data sets. It is specifically aimed at high-resolution MS data. Several labeling techniques as well as label-free quantification are supported.
› dokumaxquant:start [MaxQuant documentation] MaxQuant is a proteomics software package designed for analyzing large mass-spectrometric data sets. It is specifically aimed at high-resolution MS data. Several labeling techniques as well as label-free quantification are supported. MaxQuant is freely available and can be downloaded from this site. The download includes the search engine ...
MaxQuant | Max Planck Institute of Biochemistry MaxQuant. MaxQuant is a quantitative proteomics software package designed for analyzing large-scale mass-spectrometric data sets. It supports all main labeling techniques like SILAC, Di-methyl, TMT and iTRAQ as well as label-free quantification. Also measured spectra of various vendors - Thermo Fisher Scientific, Bruker Daltonics, AB Sciex and ...
PDF Proteome-wide label-free quantification with MaxQuant - BSPR Proteome-wide label-free quantification with MaxQuant Jürgen Cox Max Planck Institute of Biochemistry July 2011 MaxQuant Feature detection Initial Andromeda search Recalibration Main Andromeda search Consolidation / protein quantification MaxQuant Data acquisition Statistics & systems biology Raw data MQ output tables
MaxQuant error at label free normalization step - Google Groups Then no issue with maxquant search. Good luck ... I am having a similar problem, but under my description it just says the file folder and "label free_normalization 0 Label free_normalization" and then it cuts off. I'm searching 180 files and it keeps happening. All files open fine in xcalibur, and most have searched fine in smaller searches.
Comparative evaluation of label-free quantification strategies The selection of a data processing method for use in mass spectrometry-based label-free proteome quantification contributes significantly to its accuracy and precision. In this study, we comprehensively evaluated 7 commonly-used label-free quantification methods (MaxQuant-Spectrum count, MaxQuant-iB …
MQSS 2019 | L4: Label free quantification | Christoph Wichmann To address the drawbacks and make use of the advantages of the label-free approach, MaxQuant contains a label-free quantification module called MaxLFQ. It is a series of algorithms that are based...



















Post a Comment for "43 maxquant label free quantification"